Spatial Multiomics Core is a new branch of COBRE Genomics, Bioinformatics and Spatial Multiomics Cores –Tulane Center For Aging. Spatial Multiomics Core is responsible for running 10X Visium spatial slide. The Visium Spatial Gene Expression Solution measures total mRNA in intact tissue sections and maps the location(s) where gene activity is occurring. Each Visium Spatial Gene Expression Slide contains Capture Areas with gene expression spots that include primers required for capture and priming of poly-adenylated mRNA. Tissue sections placed on these Capture Areas are fixed and stained All the cDNA generated from mRNA captured by primers on a specific spot share a common Spatial Barcode. Libraries are generated from the cDNA and sequenced and the Spatial Barcodes are used to associate the reads back to the tissue section images for spatial gene expression mapping.
Why spatial transcriptomics? Because position matters!
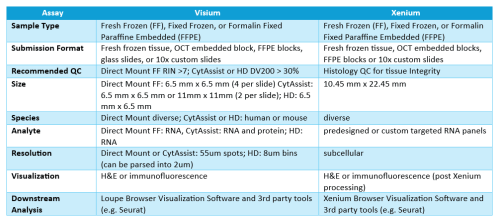
Tulane Spatial Multiomics core is a new branch of the Centers of Biomedical Research Excellence (COBRE) cores at Tulane center for aging. This core is responsible for running all the techniques and experiments for 10x genomics Visium, Visium for HD, Visium for CytAssist and Xenium in situ. Spatial transcriptomics is important because it allows researchers to study gene expression within the context of tissue architecture and organization. Here are several reasons why it is considered significant:
Preserves Spatial Information
Traditional transcriptomics techniques analyze gene expression from homogenized tissue samples, losing spatial context. Spatial transcriptomics preserves this context by linking gene expression data with the exact location within the tissue where the RNA was originally found.
Reveals Cellular Heterogeneity
Tissues are composed of diverse cell types organized in specific spatial patterns. Spatial transcriptomics helps in understanding which genes are expressed in which cell types and where, revealing the heterogeneity and functional specialization of cells within tissues.
Identifies Biomarker Signatures
By mapping gene expression profiles to spatial locations, spatial transcriptomics can identify spatially localized biomarkers associated with disease states or specific biological processes. This information is crucial for developing targeted therapies and diagnostics.
Maps Developmental Processes
It provides insights into how gene expression patterns change during development or in response to stimuli within the native tissue environment. This helps in understanding the molecular mechanisms underlying tissue formation, maintenance, and regeneration.
Advances Disease Research
Spatial transcriptomics can elucidate how gene expression changes in diseases such as cancer or neurodegenerative disorders are spatially distributed within tissues. This spatial understanding can lead to the discovery of new therapeutic targets and strategies.
Improves Drug Development
By linking molecular information to spatial tissue organization, spatial transcriptomics can aid in the development and testing of new drugs by providing a more comprehensive understanding of how drugs affect specific cell types and regions within tissues. Overall, spatial transcriptomics bridges the gap between genomic information and tissue architecture, offering a more nuanced and detailed understanding of biological systems in health and disease. This technology is increasingly important in advancing biomedical research and personalized medicine.
Xenium In situ assays RNA in Fresh frozen (FF), Fixed Fresh Frozen (FxF), or formalin fixed paraffine embedded (FFPE) tissues.
Xenium Capabilities
• Highly specific padlock chemistry for probes
• Customizable probe assays
• Free software for analysis and visualization
• High performance in situ data in days (not weeks)
• Non-destructive process compatible with IF, H&E, & 10x Genomics Visium
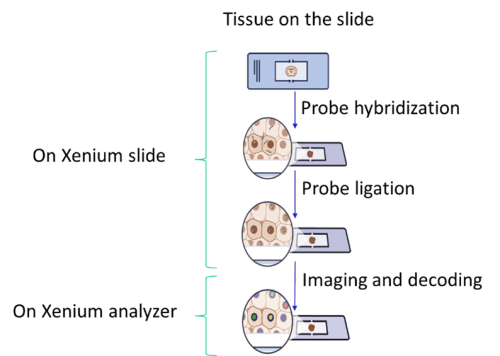
Xenium Workflow
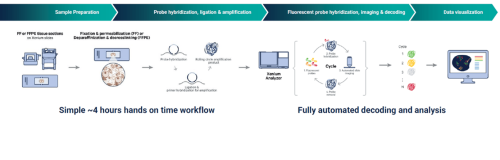
Sample Requirements
Fresh Frozen (FF), Fixed Fresh Frozen (FxF) or Formalin Fixed Paraffine Embedded (FFPE)
Section Thickness
• 5 µm (FFPE)
• 10 µm-20 µm (Fresh/Frozen; Fixed Frozen)
Sample Submission Options
Spatial Multiomics Core at Tulane center for Aging accept the following samples forms:
• Fresh frozen tissue (We do the OCT embedding)
• OCT embedded tissue blocks
• FFPE blocks
• 10X Genomics Xenium Slide with the tissue mounted.
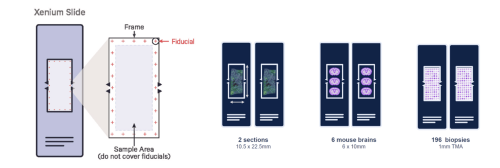
Xenium Slide Capture Area
Xenium slides have a wide imageable area outlined by a fiducial frame measuring 12 mm x 24 mm. Specimens need to fit within the available sample positioning area measuring 235 mm2 (10.45 mm x 22.45 mm). Tissue must not cover the fiducial frame.
Xenium Panels
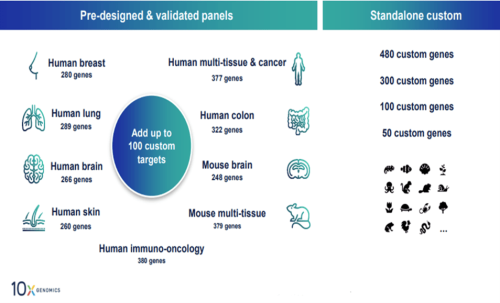
Available Pre-designed Panels:
• Human Brain (266 genes)
• Human Breast (280 genes)
• Human Colon (322 genes)
• Human Immuno-Oncology (380 genes)
• Human Lung (289 genes)
• Human Multi-Tissue and Cancer (377 genes)
• Human Skin (260 genes)
• 5K Human Pan Tissue & Pathways Panel (5K genes)
• Mouse Brain (248 genes)
• Mouse Tissue Atlas (379 genes)
• 5K Mouse Pan Tissue & Pathways Panel (5K genes)
Standalone Fully or Custom panels – you can fully customize all genes in the panel for up to 480 genes.
Add on Custom up to 100 custom targets can be added to pre-designed panels.
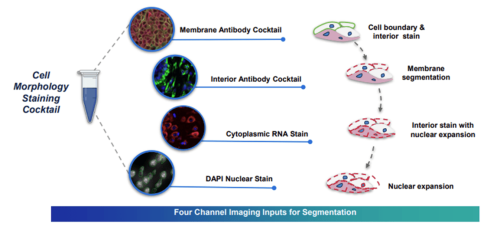
Xenium Cell Segmentation Kit
The Xenium with Cell Segmentation Staining method uses a specialized stain mix to precisely identify cell boundaries. These labels facilitate automatic cell segmentation, providing clear boundary definition when membrane signals are pronounced and alternative strategies for boundary identification when membrane staining is unclear.
The Xenium Multi-Tissue Stain Mix includes four labeling agents targeting cell membranes, cytoplasm, ribosomal RNA, and nuclei with DAPI.
Multimodal Cell Segmentation Antibodies:
Membrane markers: E-cadherin (Cdh1), ATPase1a1, CD45
Cytoplasmic RNA: 18s
Cytoplasmic proteins: vimentin, αSMA
Cell Segmentation is automatically included with 5K Pan Tissue & Pathways Panels and can be added to any other Xenium pre-designed or custom panel by request.
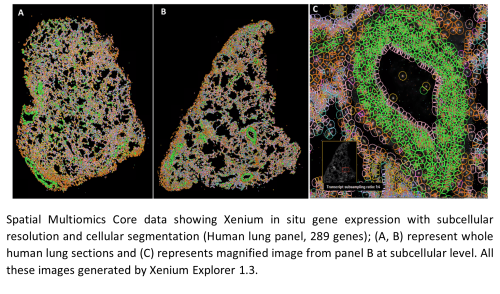
Spatial Multiomics Core Results for Xenium

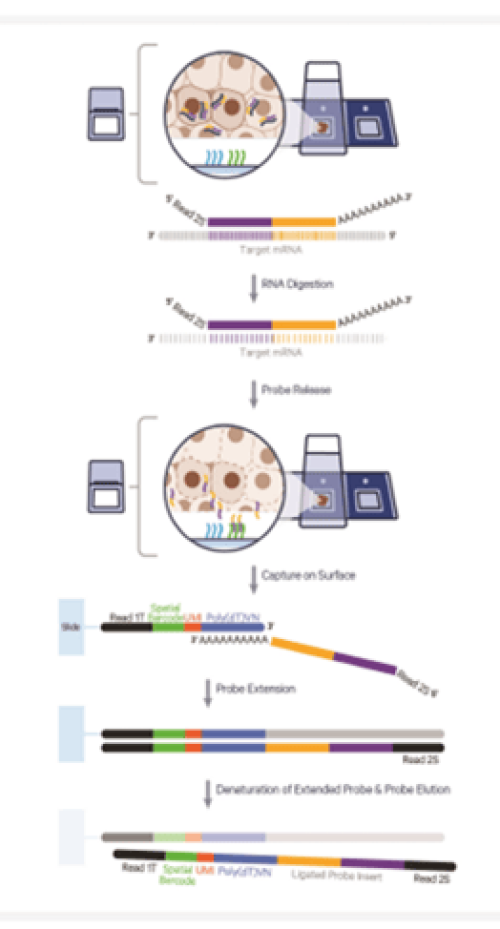
Visium for CytAssist allow automated transfer of probes from a regular histology slide or archived slides to a Visium CytAssist slide for better recovery of RNA transcripts.
Visium for CytAssist Slides
Visium for CytAssist slide come in two forms of capture areas:
Two of 6.5mm x 6.5mm capture areas have about ~5000 barcoded spots.
Two 11mm x 11mm capture area and have about ~14000 barcoded spots.
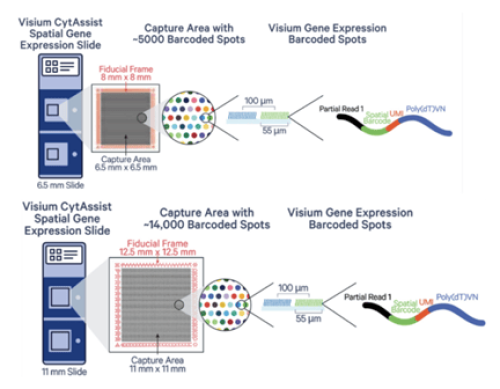
Resolution
Depending on the tissue, an average of 1-10 cells will cover a spot.
Sample Submission Options
Spatial Multiomics Core at Tulane center for Aging accept the following samples forms:
• Fresh frozen tissue (We do the OCT embedding)
• OCT embedded tissue blocks
• FFPE blocks
• Tissue mounted on a regular slide (tissue on the slide must be placed on allowable area)
• Archived slide or TMA
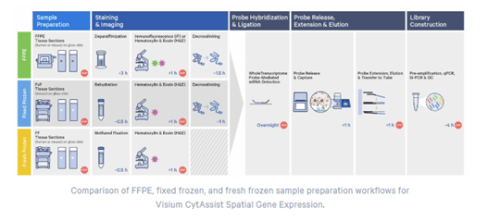
Sample Requirements
• Fresh Frozen (FF), Fixed Fresh Frozen (FxF) or Formalin Fixed Paraffine Embedded (FFPE)
• Currently supports human and mouse samples only.
• Ensure desired tissue area will fit within the 6.5 x 6.5 mm or 11 x 11 mm capture area.
• Section tissue within the allowable target area on the plain glass slide will be only transferred to the Visium CytAssist slide
Section Thickness
• 3-5 µm (FFPE)
• 10 µm-20 µm (Fresh/Frozen; Fixed Frozen)
QC for Visium for CytAssist
RNA quality of the block should be assessed prior to sectioning.
• For FF or FxF RIN should be ≥4
• For FFPE DV200 should be ≥ 30%.
HD slides contain two 6.5 x 6.5 mm Capture Areas with a continuous lawn of oligonucleotides arrayed in millions of 2 x 2 µm barcoded squares without gaps, achieving single cell–scale spatial resolution. The data is output at 2 µm, as well as multiple bin sizes. The 8 x 8 µm bin is the recommended starting point for visualization and analysis. Visium HD use the CytAssist workflow.

Resolution
Visium HD provide assay the whole transcriptome at single cell resolution.
Sample Requirements
• Currently supports Formalin Fixed Paraffine Embedded (FFPE) tissue.
• Currently supports human and mouse samples only.
• Ensure desired tissue area will fit within the 6.5 x 6.5 mm capture area.
• Section tissue within the allowable target area on the plain glass slide will be only transferred to the Visium CytAssist slide
Section Thickness
3-5 µm (FFPE)
QC for Visium for CytAssist
RNA quality of the block should be assessed prior to sectioning.
• For FFPE DV200 should be ≥ 30%.
Visium for Fresh Frozen tissue (Direct Mount Assay)
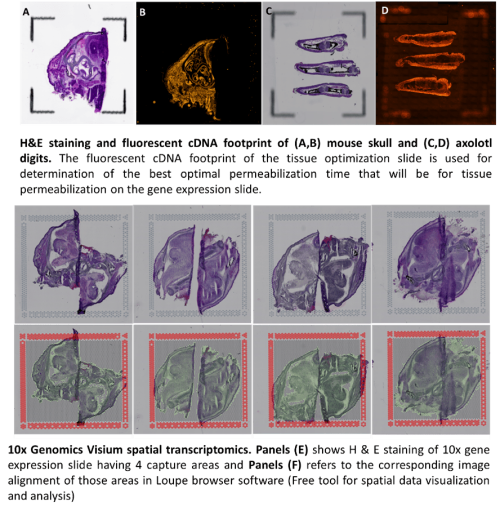
It assays the whole transcriptome in any species. Visium for fresh frozen tissue comes with two different slides with different numbers of capture areas. Tissue Optimization slide (TO slide): it comes with 8 captures areas of 6.5x6.5mm capture squares surrounded by itched frames. The purpose of TO slide is to determine the optimal permeabilization time that will be used for gene expression assay. Also, it ensure the tissue compatibility with the assay. it is not spatially barcoded slide, so it does not generate spatially barcoded library. Gene expression slide (GEX slide): it comes with 4 captures area with 6.5x6.5mm capture squares surrounded by 8x8mm fiducial frame. It is used to generate spatially barcoded library.
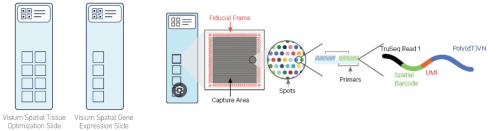
Resolution
Each capture area has about ~5000 barcoded spots. Each spot with 55µm diameter and separated from the adjacent spot by 100µm from center to center. Depending on the tissue type , an average of 1-10 cells will cover a spot.
Sample Requirements
• Freshly obtained tissue should be snap frozen to prevent RNA degradation.
• The recommended freezing method uses an isopentane and liquid nitrogen bath.
• Frozen tissue should be embedded in Optimal Cutting Temperature (OCT), a freezing and embedding compound (can be done simultaneously with freezing process)
QC for Visium for Fresh Frozen
RNA quality of the tissue block should be assessed prior to sectioning. The RNA Integrity Number (RIN) should be ≥ 7. Each new tissue type for a lab is required to undergo a tissue optimization step so that the correct permeabilization time can be established prior to scheduling a Visium GEX appointment.
Spatial Multiomics Core Results for Visium Fresh Frozen