The core can also help project investigators with:
- Refinement of experimental design, including sample size & power.
- Determination of adequate control groups, etc.
- Development (and implementation) of appropriate analytical plans to test hypotheses derived from the specific aims of each project.
- Interpretation and presentation of results from analyses.
Investigators can discuss applications and obtain information about the timeframe for services.
The Genomics, Bioinformatics, and Spatial Multiomics Core at the Tulane Center for Aging provides the following services:
We are pleased to offer full-service capabilities for spatial transcriptomics, including sequencing and bioinformatics analysis, using the 10X Visium Spatial Gene Expression and Illumina NextSeq 2000 platforms.
The 10X Genomics Spatial Gene Expression slides each contain 4 capture areas (6.5 x 6.5 mm) each with approximately 5,000 barcoded spots, containing millions of spatially barcoded capture oligomers.
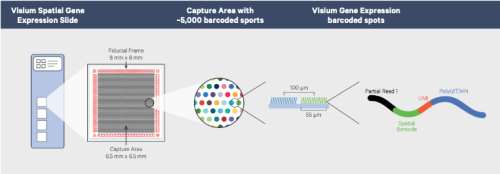
(Image source: 10X Genomics)
Tissue sections placed within the capture areas are permeabilized, and the released mRNA binds to these capture oligos, enabling gene expression information from each spot to be obtained and mapped back to corresponding areas on the tissue sections. We are currently offering 10X Visium for fresh frozen tissues; please check back for information on expanding our offerings to included formalin-fixed paraffin-embedded (FFPE) tissues.
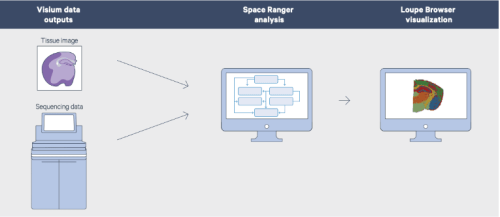
(Image source: 10X Genomics)
Please contact the Spatial Multiomics core (spatial@tulane.edu) for more information about 10X Spatial Gene Expression. We are a full-service core, and will work with you to understand your experiment, develop cost estimates and project timelines, provide expert guidance on sample preparation, perform all sectioning steps under your supervision to ensure that the correct regions of your samples are tested, and communicate with you closely at every step of the process.
The schematic below illustrates the 3 major steps in the 10X Visium spatial gene expression experiment. After completion of the first step of the protocol, 10X Visium Spatial Expression slide images and cDNA may be returned to you, or, for your ease and convenience, may be transferred to our in-house experts in the Genomics Core for library preparation and sequencing. We also are pleased to offer in-house data analysis of your Visium experimental data via the Bioinformatics Core.
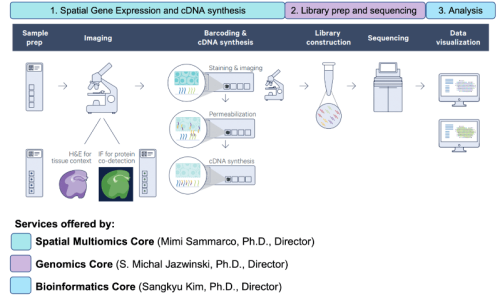
(Image source: 10X Genomics)
We are excited to announce that the Illumina NextSeq 2000 and Chromium Controller from 10X Genomics are the newest additions to our Genomics Core. We are now offering Total RNA-Seq service from library preparation through sequencing and data analysis. We also provide library preparation and sequencing services for Visium spatial gene expression slides of fresh, frozen, and FFPE samples generated from another branch of our core- Spatial Multiomics Core. Other applications such as mRNA-seq, ChIP-seq, ATAC-seq, and their single-celled counterparts are also in the pipeline. We have a highly efficient bioinformatics team that will analyze the final output data for all applications.

The Illumina NextSeq 2000 platform is a flexible next-generation sequencing platform, offering access to exome, mRNA, and single-cell RNA sequencing applications. The Illumina NextSeq 2000 platform is also used for sequencing tasks in 10X Visium spatial gene expression experiments.
Three different types of cartridges are available. They are P1, P2, and P3, producing 100 million, 400 million, or 1.2 billion single reads (or read pairs) per run respectively. Each cartridge includes an additional 38 cycles of reagent except the P3 300 cycle kit, which has 27 extra cycles.
P1 is available only as a 300-cycle kit.
P2 is available as 100, 200 or 300 cycle kits
P3 is available as 50, 100, 200 or 300 cycle kits
Sample multiplexing is available, depending on the application.
10X Genomics Chromium Controller

The Chromium Controller (Fig. 2) uses advanced microfluidics to perform single cell partitioning and barcoding which can capture molecular readouts of cell activity in multiple dimensions, including gene expression, cell surface proteins, immune clonotype, antigen specificity, and chromatin accessibility. It is compatible with Chromium Single Cell products, including Single Cell Gene Expression, Single Cell Immune Profiling, and Single Cell ATAC.
- Process 1-8 samples per run
- Collect 500 – 10,000 cells per sample
- Libraries produced are compatible with our Illumina NextSeq 2000
Sanger Sequencing: SeqStudio Genetic Analyzer

The SeqStudio is a 4 capillary electrophoresis instrument for standard sequencing with read lengths up to 900 bp. This system is useful for a wide variety of sequencing applications: SNP and STR screening, mutation detection, etc.
For sanger sequencing, we charge $5/sample.
For Fragment analysis, unit price is $4.2 or $90 whichever is high in case we prepare plate. In case of ready to run plate, unit price is $2.25 or $45 whichever is high.
Human Cell Line Authentication Service
Misidentification and cross-contamination of cell lines is a recurrent problem in life sciences research. In response, various scientific journals and funding agencies are enforcing strict usage of well-identified cell lines for research purposes and that cell line authentication must be performed prior to grant approval and/or publication. We offer comprehensive Human Cell Line Authentication Services using Short Tandem Repeat Profiling (STR) technology with AmpFLSTR Identifiler Plus PCR Kit (Life Technologies). This kit uses 16 markers in the human genome (D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, Amelogenin, D5S818, and FGA) to ensure the correct identity of your cells, avoiding the risk of misidentified or contaminated cell lines in your research.
LabChip GX Touch Nucleic Acid Analyzer

The LabChip GX Touch Nucleic Acid Analyzer (Fig. 4) provides a comprehensive solution for genomic sizing and quantitation which utilizes microfluidic technology to generate high-resolution data required for NGS library preparation (smear and fragment analysis) and quality control, RNA and DNA fragment analysis etc.

The Agilent 2100 Bioanalyzer (Fig. 6) provides highly reproducible on-chip electrophoresis that allows sizing, quantitation and quality control of DNA and RNA. It is used extensively in the NGS workflow for quality control and provides quality determination of RNA samples prior to RNA-seq.
QuantStudio 7 Flex Real-Time PCR System

The Applied Biosystems™ QuantStudio™ 7 Flex Real-Time PCR System (Fig. 6) delivers the proven reliability, sensitivity, and accuracy of the data in a new industrial design. The QuantStudio 7 Flex System has been optimized to enable the broadest range of quantitative PCR applications, with additional dyes, formats, and automation options.
Location and Contact Information
Genomics, Bioinformatics, and Spatial Multiomics Core
School of Medicine
6th floor
Room 6304
1430 Tulane Avenue
New Orleans, LA 70112
Phone: 504-988-4238
E-mail: mawoal@tulane.edu
Core-specific contacts for more information
Spatial Multiomics Core: Ahmed Moustafa and Quincy Brown (spatial@tulane.edu)
Genomics Core: Muhammad Awoal and Michal Jazwinski (genomicscoreaging@tulane.edu)
Bioinformatics Core: Sangkyu Kim and Chenyao Xiao (skim5@tulane.edu)
General questions, quotations, invoicing: Quincy Brown (jqbrown@tulane.edu)
Acknowledgements
The COBRE Genomics, Bioinformatics, and Spatial Multiomics Core at the Tulane Center for Aging, Tulane School of Medicine is supported by NIH: NIGSM COBRE grant P20GM103629 and P30GM145498 to Dr. S. Michal Jazwinski.
The Ion Chef was funded in 2016 by Core Lab Equipment Award from the Tulane School of Medicine.
