Tulane Pathology Spatial Genomics Center (TPSGC) offers Visium CytAssist spatial transcriptomics service to the research community at Tulane.
Conventional Bulk RNA-sequencing analyzes the RNA mixtures and determines the average expression for each gene across all the cells within the tissue. Unfortunately, the spatial information is inevitably lost during tissue dissociation. The new Visium technology enables us to conduct complete transcriptomic analyses of fixed and frozen tissue segments without sacrificing spatial information. Thus, gene expression data can be assessed in connection with tissue structure, tissue microenvironments, and cell clusters. Visium is done via RNA templated ligation of pairs of gene target probes, which is a more sensitive and specific technique than other methods such as targeted amplicon sequencing or sequence-specific hybridization. Further, this method reduces the bias that can be introduced by the latter.
TPSGC has recently implemented the Visium CytAssist platform for spatial transcriptomic analysis which can be used to analyze both human and mouse tissues, in healthy and diseased states, for numerous applications.
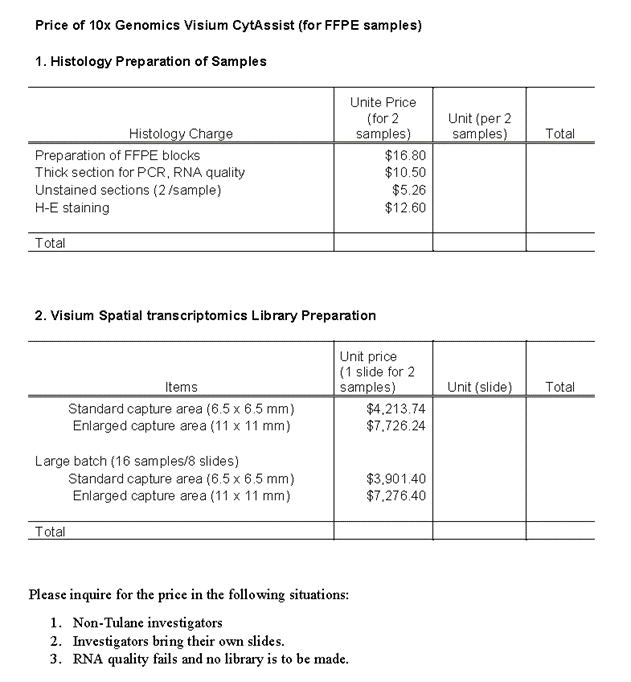
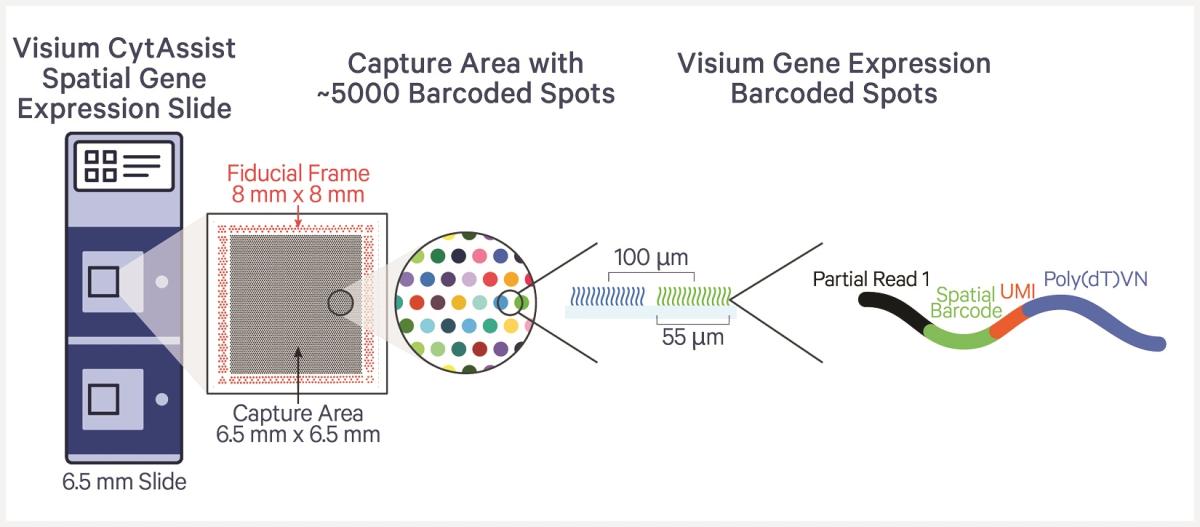
The Visium CytAssist platform requires the use of Visium slides, each of which is comprised of two Capture Areas. Every Capture Area has ~5,000 capture spots, each with millions of oligonucleotides that are made up of a 30 nucleotide poly(dT) sequence to capture polyadenylated mRNA molecules, a 12 nucleotide unique molecular identifier (UMI) to identify any duplicates that may arise during library preparation and sequencing, a 16 nucleotide Spatial Barcode that is common to all oligonucleotides in each gene expression capture spot, and a partial TruSeq Read 1 sequence for use during library preparation and sequencing.
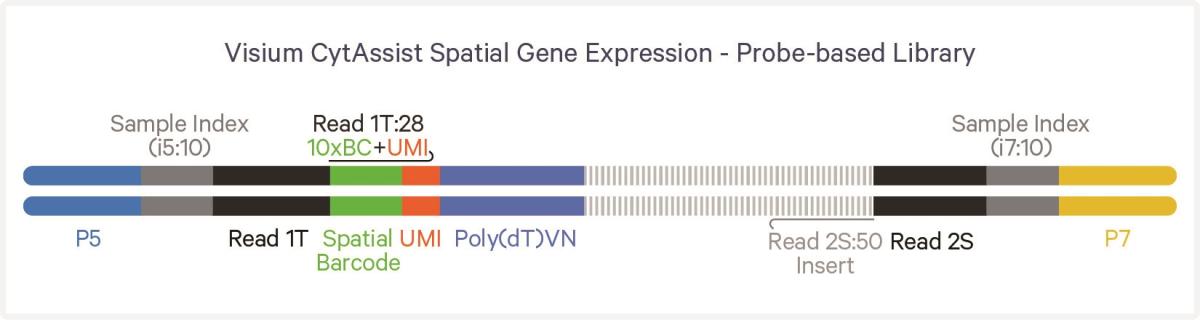
The Visium CytAssist instrument will be used to transfer transcriptomic analytes from tissue samples on standard glass slides to Visium slides. It is compatible with both fresh frozen and FFPE tissue sections and eliminates the need for directly sectioning onto Visium slides, thus allowing for more streamlined sample preparation and the gathering of spatial profiling insights from more samples.
In the Visium CytAssist pipeline, sectioning, deparaffinization, staining, and imaging (H&E or IF) are carried out on standard glass slides. After probe hybridization (Step 3), two standard slides and two Capture Area Visium slides are placed in the CytAssist instrument, with the tissue sections on the standard slides aligned over the Capture Areas. Inside the instrument, a brightfield image is taken for data analysis, followed by hybridization of transcriptomic analytes to the Visium slide (Step 4). The rest of the steps, commencing with probe extension, are performed outside the CytAssist instrument (Steps 5–6).
The Visium CytAssist instrument will be used to transfer transcriptomic analytes from tissue samples on standard glass slides to Visium slides. It is compatible with H&E or IF-stained FFPE (and frozen) tissue sections and eliminates the need for directly sectioning onto Visium slides, thus allowing for more streamlined sample preparation and the gathering of spatial profiling insights from more samples.


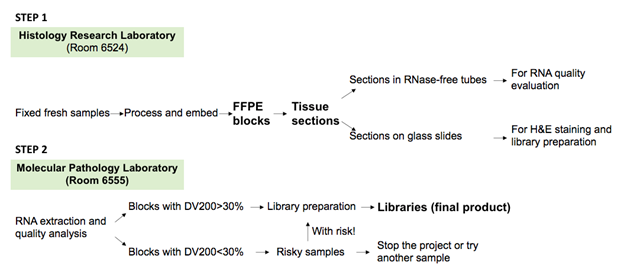
TPSGC is located on the 6th floor of Hutchinson Building, Room 6555 and 6524 in the Department of Pathology, Tulane University School of Medicine.
Visium CytAssist libraries can be sequenced with paired-end dual indexing. Sequencing data can be analyzed using the SpaceRanger software hosted on the Tulane Cypress High Performance Computing System. SpaceRanger software will process both the imaging data and sequencing data simultaneously and generate the spatial gene expression matrix which can be further processed by spatial transcriptomic analysis software such as Seurat.
For sequencing by the facility at Tulane School Medicine, please contact:
Tulane Center for Translational Research in Infection & Inflammation ~ NextGen Sequencing Core
Di Tian, MD, PhD
Dtian2@tulane.edu
Director, Histology Laboratory and Molecular Pathology Laboratory
Department of Pathology and Laboratory Medicine
For more information
Zhen Lin, MD, PhD
zlin@tulane.edu
Co-Technical Director, Spatial Genomics Services
Department of Pathology and Laboratory Medicine
Shengmin Yan, PhD
Syan2@tulane.edu
Co-Technical Director, Spatial Genomics Services
Department of Pathology and Laboratory Medicine
Xiaojiang Xu, PhD
Xxu17@tulane.edu
Bioinformatics and Genomics Analysis Core
Department of Pathology and Laboratory Medicine
Tyler Rorison, HTL(ASCP)CM
TRorison@tulane.edu
Manager, Histology Laboratory
Department of Pathology and Laboratory Medicine