Wimley Lab
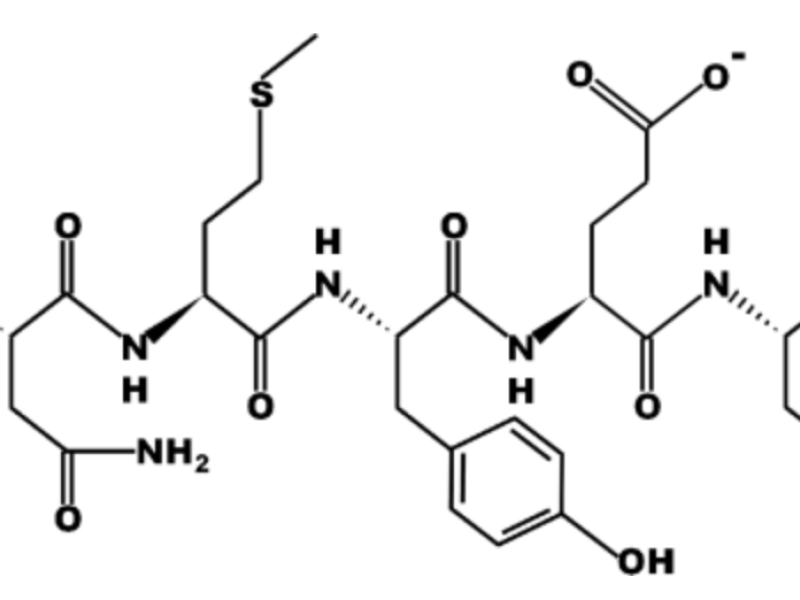
Welcome to the Wimley Lab at Tulane University School of Medicine! Our lab explores how proteins are structured, folded, and designed within membranes. By using peptide models of membrane proteins, the team develops new ways to understand and engineer these essential biological systems.
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Publications
The Wimley Lab is internationally recognized for advancing our understanding of how peptides and proteins interact with cell membranes, with research spanning fundamental biophysics to translational medicine. Using synthetic molecular evolution, combinatorial design, and high-throughput screening, the team has developed novel antimicrobial peptides, viral entry inhibitors, and peptide-based drug delivery systems with strong therapeutic potential. Their work has been featured in top journals such as Journal of the American Chemical Society, ACS Chemical Biology, Journal of Biological Chemistry, and Biophysical Journal, establishing the lab as a leader in peptide engineering and membrane biology.
The Wimley Lab is internationally recognized for advancing our understanding of how peptides and proteins interact with cell membranes, with research spanning fundamental biophysics to translational medicine. Using synthetic molecular evolution, combinatorial design, and high-throughput screening, the team has developed novel antimicrobial peptides, viral entry inhibitors, and peptide-based drug delivery systems with strong therapeutic potential. Their work has been featured in top journals such as Journal of the American Chemical Society, ACS Chemical Biology, Journal of Biological Chemistry, and Biophysical Journal, establishing the lab as a leader in peptide engineering and membrane biology.

News

Contact Us
Wimley Lab
Tulane University School of Medicine
Biochemistry and Molecular Biology, 8643
1430 Tulane Avenue
New Orleans, LA 70112-2699
Rm 6032
phone: 504-988-7076